Inducing motility of escherichia coli cells
[2023: Explanations still to be write]
Preliminary report, February 19, 2014
Viafx24
Institut Curie, UMR168, 26 Rue d'Ulm, Paris, France
Introduction
In these preliminary experiments, we have tried to control motility of Escherichia coli cells. A cyaA mutant can't produce flagellum and thus can't swim. In this mutant, adenylate cyclase, the enzyme encoded by the cyaA gene, is absent and thus cells can't synthesize cAMP. Yet, cAMP is needed to activate the transcriptional factor CRP that allows expression of the operons of motility. To sum up, without cyaA, there is no production of cAMP, no activation of CRP, no expression of genes of motility, no flagellum and thus no swimming. However, if one adds cAMP to the medium, cyaA mutant is "complemented" and cells should recover the capability to swim. In the following experiment, we used a double mutant cyaA- cpdA-. This latter deletion removes the phosphodiesterase responsible for degradation of cAMP. This allows to activate synthesis of flagellum with much lower cAMP concentrations.
Materials and methods
Culture and slide preparation
Precultures were performed in M9 medium with 0.4% (w/v) glucose and 0.1%(w/v) casamino acids. We inoculated the medium from a frozen stock of the strain called cyaA- cpdA- p-sdh-gfp and let cells grew overnight at 37°C under shaking. Then we mixed 20μl of this preculture with 180μl of fresh identical medium and added the indicated concentration of cAMP from a stock at 1mM. We placed a thin empty square of PDMS on a slide to create a chamber and added 28μl of the above medium. Then, the chamber was closed with a cover-slip. This is time 0.
Acquisition
The slide is placed under a 50X objective of the Reichert-Jung Polyvar Met microscope. The Z is approximately placed between the slide and the coverslip. The X and Y are placed closed to a border in order to optimize oxygen access. Acquisition is performed with the photometric evolve 512 camera (512px/512px). Exposition time was set at 10ms. The total acquisition time between frames was 60ms (experiment 1) or 40ms (experiment 2). We used custom MATLAB scripts to acquire the images (using micromanager java classes). We acquired 500 images approximately each 5 minutes during several hours (105 images).
Particles and trajectories detection, data and images processing
We used the particle tracker plugin of ImageJ to detect and track individual cells. We implemented MATLAB scripts that automate the process (using the MIJI adapter from EPFL). Briefly, the script opens 200 images, detects trajectories, computes the mean square displacement, saves the data and passes to the next batch of images (next time point). This script is time consuming: several hours to get all the data. We also wrote some scripts that measure several parameters such as speed of the cells or scripts that clean images to focus on particular trajectories.
Results
Recovery of motility over time after adding cAMP to the medium
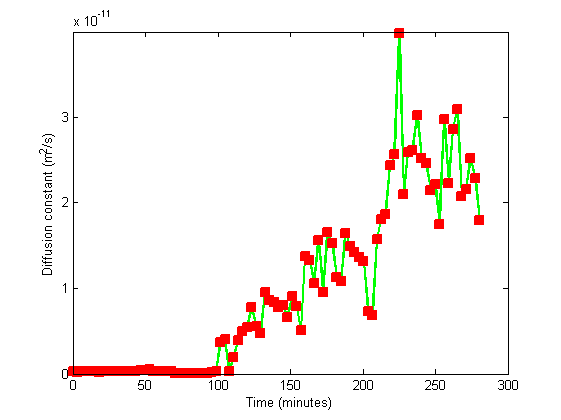
One can observe that cells start to decant (they leave the field) given that they can't swim. They come back after recovery of motility, as expected.
Recovery of motility function of cAMP concentrations
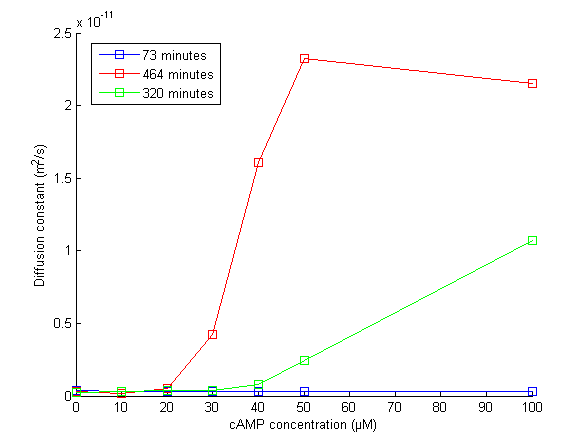
On movie 2, one can observe the rapid recovery of motility when using the strongest concentrations of cAMP (50μM and 100μM) compare to the others. However, on can notice that after 1000 minutes, all the concentrations (except the control: 0μM not shown) recovered motility. However, by changing the Z and observing the bacterial layer on the slide where cells have decanted (end of the movie), one can observe significant differences between the concentrations: for lower concentration, the layer is very dense. This means that only few cells recovered motility. At the opposite, for strongest concentrations, there is few cells deposited on the slide. This means that most of cells are swimming in the medium. This can be considered as a bistable phenotype. In other word, by ranging the cAMP concentration, one may control the proportion of cells that are going to recover motility (probably due to stochasticity in gene expression).
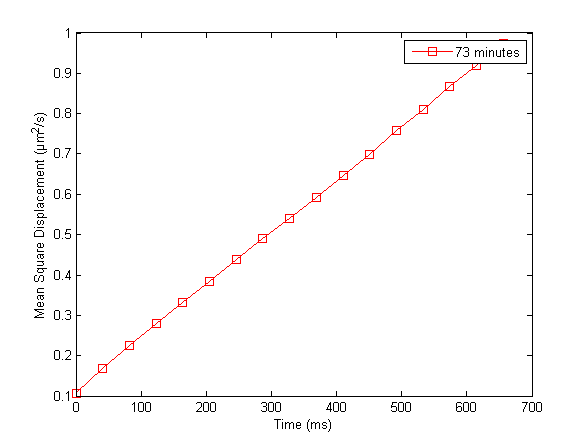
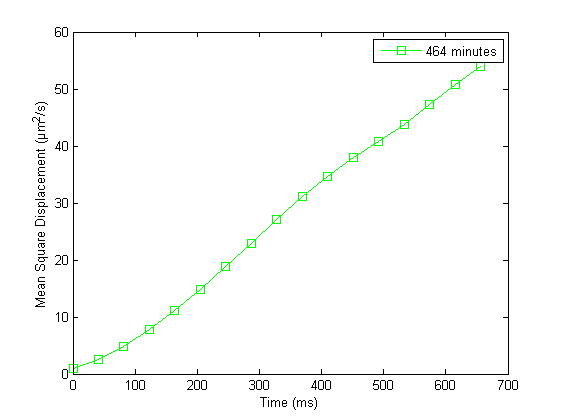
Discussion
Biology
We succeeded to control the induction of motility by using cyaA- cpdA- cells. The main experiment has been reproduced at least five times and led to similar results: after about 2 hours in presence of exogenous cAMP, we observed the recovery of motility. The difference of motility between cells that swim and cells that don't swim is very high (factor 50-100). The experiments were performed at room temperature. Quicker induction may be possible at 37°C since gene expression would be faster. Rapidity of the induction also depends of the state of the cells at the beginning of the experiment: stationary phase cells are less reactive since they need to leave the lag phase (second experiment: ~3 hours before the recovery of motility). Cells that are still in exponential phase shows a quicker induction of motility: ~100-120 minutes (first experiment). We also try to perform the opposite experiment: using already motile cells and removing cAMP of the medium by centrifugation to measure the fall of motility. Those experiments (not shown) led to ambiguous results: centrifugation seems to directly stop motility (for 99% of the cells): flagellum may be broken during this step. Without cAMP, recovery of motility never comes back and cells decant on the slide. Those kinds of cells are considered not naive. We mean that they do not swim but have ever swum: they should still have a cellular content acting as a memory of the past (swimming). When we uses not naive cells that remain in a medium without cAMP for 3 hours and adds again cAMP, the recovery of motility is faster, as expected (70-100 minutes). This may be considered as a kind of hysteresis (memory effect).
Failures
The quality of images (transmission) may be improved in order to improve trajectories detection. The best way consists to use fluorescence and thus, the corresponding adequate plasmid. We tried to use the pZE-gfp plasmid that produces a huge amount of GFP. However, the cyaA- cpdA- strain that has already a growth defect, do not seems to support this plasmid: after transformation, colonies appear very slowly, cells are fluorescent but look sick and they never recover motility after addition of cAMP to the medium. I tested several other plasmids from my collection without success: weaker promoters allow healthy cells but the fluorescence signal is too low to provide interesting images. Stronger promoters leads to the same effect that the pZE-gfp plasmid. Nevertheless, I am still confident to find the intermediate plasmid/promoter allowing a good signal without sicken the cells.
Concerning acquisition and data processing, it remains some little problems that I do not solve:
- The Z still doesn't work and can't be automated for now. Solving this problem may allow to follow a single cell during hours and then observing the different steps before the recovery of a normal motility. In particular, I believe that cells swim in circle for a while when they begin to get the first flagellum.
- The acquisition process still has a bug (probably the camera driver). 1/10 of acquisitions will stop before the end due to a camera error.
- The script that calculates the trajectories on all the batch of images (105 images) has a limit: It can't be launched on more than 200 frames per time point due to an unsurpassable limit of MATLAB (java heap memory cannot exceed 1GB).
- The script that cleans the images in order to follow specific trajectories (cells that are present on at least X frames or cells that have a speed of at least Y) is very time consuming (one night to compute a batch of thousand images). However, parallelization of the code using parfor loop is probably feasible: a gain of about a factor 5 should be easily obtained allowing computation to be completed in the order of an hour.
Conclusion
Inducing the motility of cyaA- cpdA- cells by adding cAMP to the medium is easy to perform and perfectly reproducible. It takes approximately 2 hours at room temperature to be achieved and the optimal concentration of cAMP is closed to 100μM. Interesting phenotypes such as bistability or hysteresis may also occur and thus could be investigated in a deeper way. The scripts of acquisition, data and images processing are fully functional: they allow to acquire/ to extract information from a huge batch of images (105). Most importantly, they can be run in parallel. I mean that a feedback loop is easily implementable: scripts can automatically detect, for instance, the induction of motility and then act on the acquisition process (changing XY or increasing the frequency of acquisition of images for instance). I believe that all these tools may be useful when we will use micro fluidic devices. Those latter may help to investigate different questions of interest. To my opinion, this could be the next step.